Molecular cloning is a technique used to create DNA experimental tools that are expressed in either bacterial or mammalian cells. Molecular cloning is defined as the isolation and amplification of a target DNA sequence.

DNA. Image Credit: jijomathaidesigners/Shutterstock.com
What is Molecular Cloning?
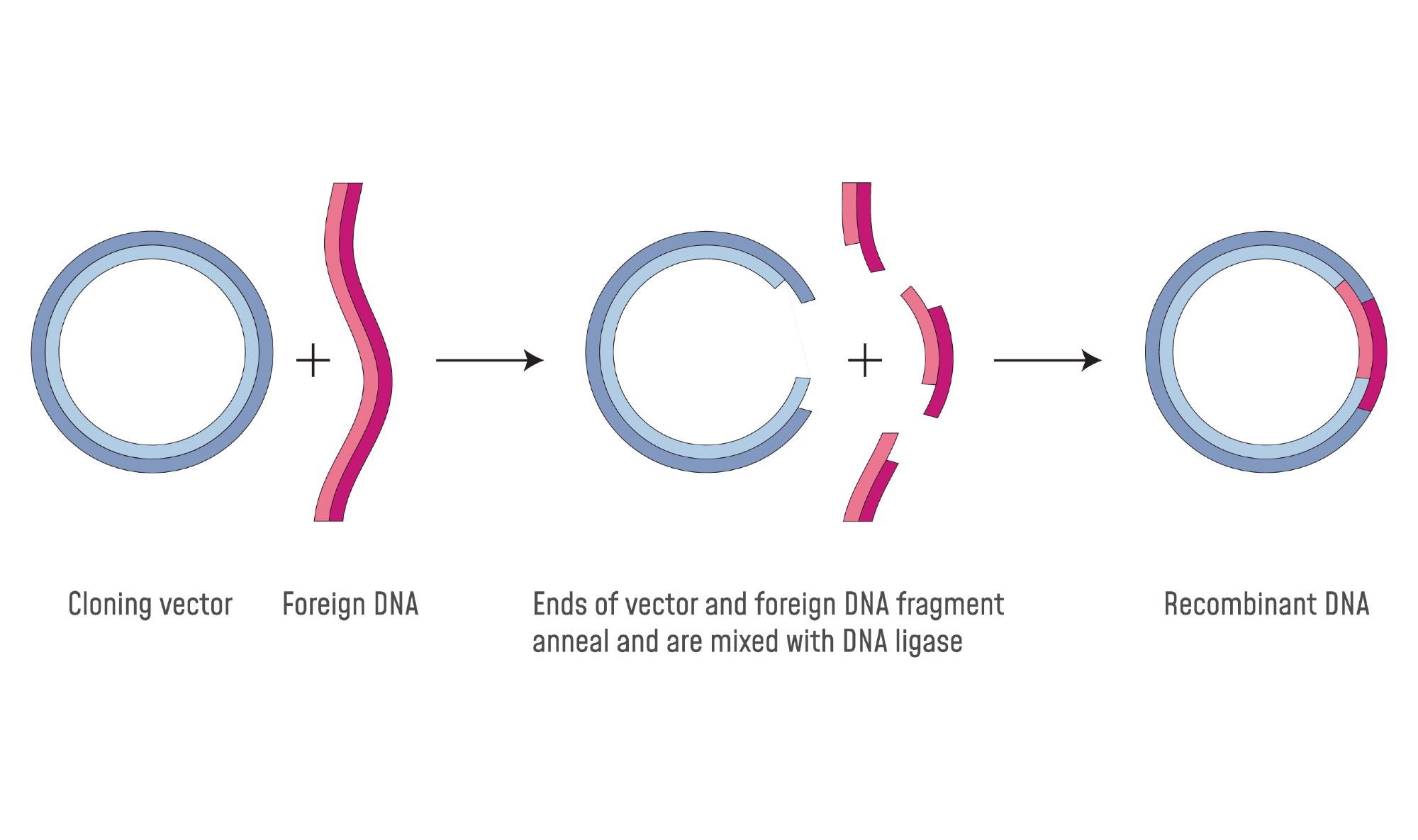
Molecular cloning is a technique that involves inserting a fragment of foreign DNA into a vector that is capable of replicating autonomously in a host cell. This host cell is usually a bacteria (Escherichia coli). The production of several copies of the inserted DNA can be used for several purposes.
The Requirements for Molecular Cloning
To conduct a cloning experiment, several materials are required. These include the target DNA, a host organism, vector DNA for cloning into, a means of inserting foreign DNA into the vector, a method of placing the in vitro modified DNA into the host cell, and methods for selecting and/or screening cells that carry the inserted foreign DNA.
Enzymes Involved in Molecular Cloning
There are three types of enzymes used for cloning. These include restriction endonucleases, polymerases, nucleases, ligases, kinases, and phosphatases. Restriction endonucleases are bacterial enzymes that cleave double-stranded DNA either at or near recognition sites in nucleic acid molecules, known as restriction sites.
The majority of the inserted fragments are produced as a consequence of digesting an existing piece of DNA with enzymes called restriction enzymes. Are a result, short inserts of ~ 100 base pairs in length are generated, these short inserts can also be synthesized commercially as single-stranded oligonucleotides which are then annealed to form a double-stranded fragment.
Polymerases are enzymes that catalyze the synthesis of DNA or RNA polymers whose sequence is complementary to the original template.
DNA nucleases catalyze the breaking or cleavage of the phosphodiester bonds that exist between deoxyribose and the phosphate residue in the DNA strand backbone. Exonucleases are nucleases that cleave at the proximity of DNA ends; those that cleave within the DNA strand are called endonucleases. These do not require a free DNA end for cleavage. Some enzymes have both activities.
Ligases form phosphodiester bonds to join two pieces of DNA; they utilize is adenosine triphosphate (ATP) in the presence of a magnesium ion. Kinases transfer phosphate groups from donor molecules, while phosphatases catalyze the removal of phosphate residues.
Molecular cloning. Image Credit: Fancy Tapis/Shutterstock.com
The Choice of Vector DNA
DNA molecules that function as molecular carriers are called vectors. They carry the DNA of interest into the host cell and facilitate its replication. There are three types of vectors used in cloning:
- Plasmids which are used to clone small segments of DNA, approximately 10 to 15 kb
- Bacteriophage gamma: used to clone larger segments of DNA (~20kb)
- Cosmids: these are plasmids containing DNA sequences from bacteriophage gamma used to clone larger fragments (up to 45kb in length)
Following isolation of the target DNA sequence, this insert is located to a vector plasmid to produce a construct the plasmid is a double-stranded circular piece of DNA that can be propagated in bacterial cells such as E. coli. In the laboratory, vectors are typically smaller versions of naturally occurring plasmids that have several features:
- A replication origin
- A reporter gene which is typically a drug-resistant gene
- Unique restriction sites that enable the DNA fragment to be easily inserted. These restriction sites are often clustered in polylinker regions or multiple cloning sites which enable the researcher to select the most convenient restriction enzyme combination for several inserts. A restriction enzyme can cut DNA at a specific target. The choice of restriction enzyme is considered to be a critical step when designing a cloning strategy. Several sever the double-stranded DNA in one place, creating blunt ends. Other forms of restriction enzymes allow for an overhang of a small number of bases at the site of the cat. These are referred to as complementary sticky ends, so-called as they can locate one another easily, which augments the efficiency of the ligation reaction. This subsequently increases the chances of a successful cloning event. A considered thought of the combinations of restriction enzymes can control the direction in which the insert is inserted and this is critical to many applications
The Molecular Cloning Strategy
There are four basic steps of molecular cloning:
- Linearization of the vector as a result of restriction digest at multiple cloning sites.
- Ligation off the desired or target piece of DNA into the bacterial plasmid: ligation is the means of inserting DNA into the vector. The requirements for ligation reaction consists of two or more fragments of DNA, either blunt or cohesive, a buffer containing ATP, and a DNA ligase
- Transformation of this construct into a vector such as E. coli: This is the method of placing the in vitro modified DNA into the host cell. In the process of transformation, bacterial cells take up naked DNA molecules. Following this the cells are made ‘competent, that is, amenable to receiving DNA period to achieve this cells are treated with ice-cold calcium chloride and subsequently heat shocked. On average there is an efficiency of 107 to 108 transformed colonies per microgram of DNA, and a maximum transformation frequency of 10-3. Transformation efficiency is defined as the number of colony-forming units (CFU) per microgram of DNA used to transform the bacteria. After this, electroporation of the DNA into the host cell occurs. This is a form of electric-field mediated membrane permeabilization; a high strength electric field is applied in the presence of DNA.
- Screening for the presence of positive ligation events: this is a method for selecting cells that carry the inserted DNA. It refers to the application of conditions that promote the growth of cells that carry the vector or vector and insert. Most commonly these screening methods include selecting for antibiotic resistance, checking nutrient requirements, using a blue-white selection using beta-galactosidase, very specific types of screening such as the use of hybridization. Screening allows cells to grow and tests the resulting clones for the presence of the insert
In a typical DNA cloning procedure, a DNA fragment of interest, most commonly a desired human protein, is inserted into a construct. These DNA constructs include a promoter element (a sequence of DNA needed to turn a gene on or off) which is attached to a reporter gene or a complementary DNA sequence that is under the control of a promoter.
A reporter gene is a gene that produces a molecule that can be easily assayed following transfection, a process that involves introducing DNA or other nucleic acids into eukaryotic cells and can be used as markers for screening successfully transfected.
In sum, molecular cloning involves the preparation of a vector, a ‘transformation’ which is any vehicle, typically a virus or a plasmid that is used as a vehicle to carry a DNA sequence into a host cell as part of a molecular cloning procedure.
Reference
- Lessard, J. (2013) Chapter Seven – Molecular Cloning. In Jon Lorsch (Ed.) Methods in Enzymology. Academic Press (pp. 85-98).
Further Reading
- All Cloning Content
- Cloning Human Cells
- Molecular Cloning
- Organism Cloning
- Cloning Unicellular Organisms
Last Updated: Mar 8, 2022

Written by
Hidaya Aliouche
Hidaya is a science communications enthusiast who has recently graduated and is embarking on a career in the science and medical copywriting. She has a B.Sc. in Biochemistry from The University of Manchester. She is passionate about writing and is particularly interested in microbiology, immunology, and biochemistry.
Source: Read Full Article