By using new methods for analyzing DNA data and medical records, researchers from Brown University are helping improve the understanding of complex traits that will make more discoveries relevant to non-white, non-European ancestry groups.
In a study published in the May 5 issue of The American Journal of Human Genetics, the researchers illustrated examples of the robust associations of trait determinants, or patterns of similarity, while studying 25 traits in over 600,000 individuals from seven diverse human ancestries. With these similarities, discoveries about the nature of diseases or illnesses and their responses to potential treatments become more relevant to larger groups of people—including populations that had previously been ignored or understudied.
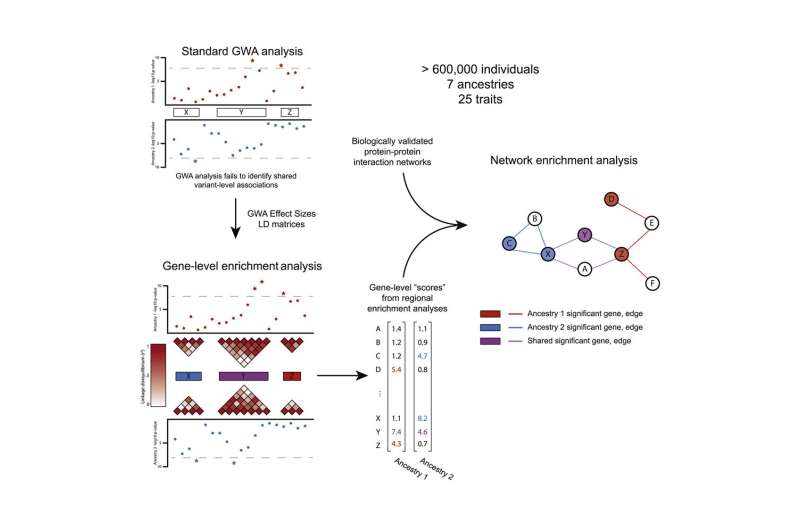
Genome-wide association (GWA) datasets, which are commonly used by geneticists, are based on the assumption that individual genetic mutations underwrite the genetic basis of traits, explained study author Sohini Ramachandran, a professor of biology and computer science who directs both the Center for Computational Molecular Biology and the Data Science Initiative at Brown. The idea is that a discovery about those mutations will be relevant to all people across a range of diverse ancestry groups, so that if the finding is used to develop treatments for genetic conditions, it will be applicable for all people with that condition.
However, recent studies have shown that GWA results estimated from self-identified European individuals are not transferable to non-European individuals. Because of this, the insights from the datasets are largely biased toward sampling individuals with European ancestry. The statistical hypothesis underlying the GWA framework is unfairly restrictive, Ramachandran said.
Thus the researchers used a new “enrichment analysis” methodology, previously developed in a collaboration between Ramachandran and Brown assistant professor of biostatistics Lorin Crawford, to address bias and underrepresentation.
“In this paper, which involved very careful analysis of a ream of data across multiple biobanks, we show that data viewed only through a very specific GWA lens may look disparate and irreconcilable,” Ramachandran said. “Yet, viewed in a more equitable way, with a more expansive methodology, it becomes biologically unified, interpretable, and, importantly, actionable.”
Ramachandran’s interest in the topic started when she learned about a study showing that children with acute lymphoblastic leukemia had different responses to the standard treatment regimen depending upon their ancestry group—for example, non-white children were more likely to relapse and had a worse prognosis. As an evolutionary biologist and population geneticist, Ramachandran started thinking about the increasing reliance on GWA studies in the development of “personalized” treatments for diseases and conditions.
“There wasn’t a lot of discussion around the extent to which the results from these studies were going to be directly applicable to all ancestries,” she said. “And based on population genetics and theory, it just seemed really unlikely that this was going to pan out in a way that was equitable.”
A fresh look at the data
With other Brown researchers, including Crawford and Samuel Pattillo Smith, Ramachandran started working on developing statistical techniques that moved beyond individual mutations to include genes and pathways.
It’s not as if this information didn’t exist; over the past two decades, funding agencies and biobanks around the world have made enormous investments to generate large-scale datasets of genotypes, exomes and whole-genome sequences from diverse human ancestries, which are then merged with medical records and quantitative trait measurements. However, the researchers explained, analyses of such datasets are usually limited to the GWA association analyses that assume a direct correlation between mutations and traits.
The researchers studied 25 traits in 566,786 individuals from seven diverse self-identified human ancestries in the U.K. Biobank and the Biobank Japan, as well as 44,348 individuals from the PAGE Consortium including cohorts of African American, Hispanic and Latin American, Native Hawaiian, and American Indian/Alaska Native individuals. They performed statistical tests of association at the mutation, gene and pathway level for the 25 quantitative traits.
They identified 1,000 gene-level associations that are genome-wide significant in at least two ancestry groups across these 25 traits, as well as pathway associations in European, East Asian and Native Hawaiian groups. A majority of these would not have been identified using GWA alone, the researchers said.
“Instead of focusing on the single mutation statistical tests—GWA—we are basically opening up a larger retinue of tests that can look for patterns at the gene level or the biologically annotated pathway level,” said Pattillo Smith, a computational biology Ph.D. candidate in Ramachandran’s lab. “For a long time, scientists have been so focused on the effect of individual mutations that a lot of valuable information is being ignored in GWA studies or going unreported in resulting publications—especially in ancestry populations that have smaller cohorts because the test at the mutation level is incredibly sensitive to a number of confounding factors. One of the benefits of aggregating across mutations to the level of a region or gene is that you can kind of smooth those things over and be more robust in your detection of the genome-to-trait relationship.”
The researchers were aiming for what’s called “biological interpretability,” Ramachandran said, “which is how we could deploy these methods in a way to analyze biobanks to their full extent, and take advantage of all the information they have to offer.”
Applying unbiased methodology to biased data sets
In the paper, the researchers discuss how biobanks are heavily skewed toward people who self-identify as having European ancestry, noted Crawford, an assistant professor of biostatistics at Brown affiliated with the Center for Computational Molecular Biology. A hidden gem of this new research, Crawford said, is that it shows how developing sophisticated statistical methods can help overcome limitations like an underrepresented sample of non-European ancestry groups.
“You don’t have to wait until the number of people from other ancestry groups is equal to the number of people self-identifying as European,” Crawford said. “In fact, even if more data is generated, the same imbalance is likely to be perpetuated. In the meantime, statistical methods at higher scales of genes and pathways can still help us gain insight into genetic architecture that can be applied in a beneficial way to these underrepresented ancestry groups. This methodology can help us use data more equitably, right now.”
In a field like genomics, the stakes are high, Ramachandran said.
Source: Read Full Article