The causative agent responsible for the unprecedented coronavirus disease 2019 (COVID-19) pandemic is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The mechanism of entry of SARS-CoV-2 involves the binding of the viral spike (S) protein to the cellular angiotensin-converting enzyme (ACE) 2 receptor that leads to proteolytic processing of the S precursor into the active S1 and S2 subunits. However, there are other factors responsible for SARS-CoV-2 entry, propagation, and pathogenesis.
In a previous study, researchers have shown that a variant of SARS-CoV-2 isolated in the Netherlands on 12 February 2020 seizes interferon-induced transmembrane (IFITM) proteins, especially IFITM2, as entry cofactors during infection.
The World Health Organization (WHO) has classified four SARS-CoV-2 variants as variants of concern (VOCs): B.1.1.7, B.1.351, P.1, and B.1.617.2. These are also known as alpha, beta, gamma, and delta variants, respectively.
In a study recently published on the bioRxiv* preprint server, researchers investigated the IFITM2 dependency of SARS-CoV-2 VOCs, including the delta variant for efficient infection.
Interferon-induced transmembrane proteins for efficient SARS-CoV-2 infection
IFITMs are a family of IFN stimulated genes (ISGs) known to protect cells against infection by many viruses, such as human immunodeficiency viruses, influenza A, rhabdo, and highly pathogenic coronaviruses, including SARS-CoV-2.
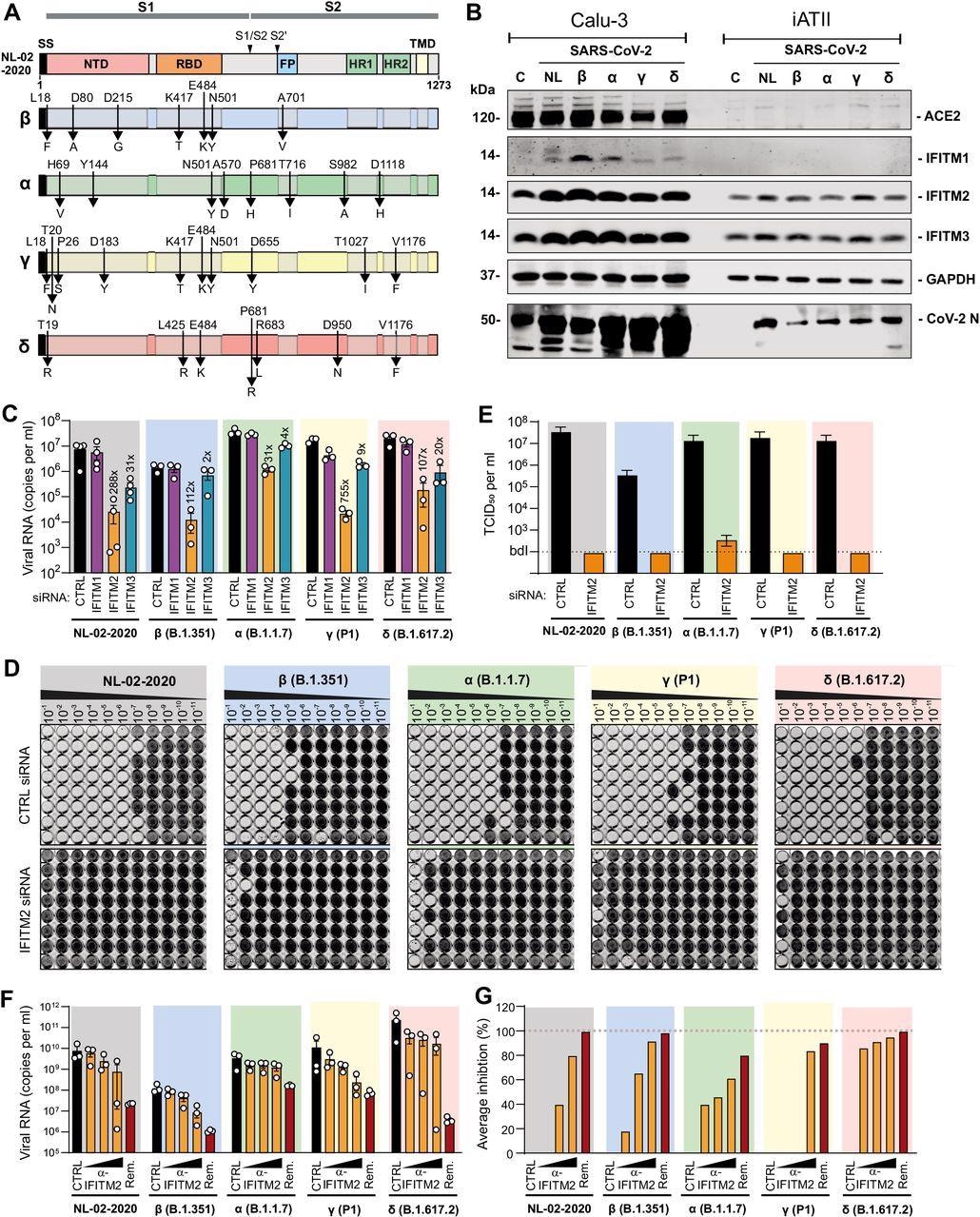
In this study, the researchers used the human epithelial lung cancer cell line Calu-3 due to its ability for siRNA knockdown (KD) of IFITM expression and iPSC-derived alveolar epithelial type II (iATII) cells as a model for the primary target cells of SARS-CoV-2 infection in the distal lung.
The Western blot analysis could not detect the ACE2 expression by iATII cells; however, it was easily detectable using fluorescence-activated cell sorting.
Outcomes of the study
The results showed that the SARS-CoV-2 Delta variant of concern in comparison to NL-02-2020 in Calu-3 cells generated 3-fold higher levels of viral RNA.
The outcomes of the study showed that there was a reduction of viral RNA production from 31- (Alpha) to 754-fold (Gamma) on depletion of endogenous IFITM2 expression in Calu-3 cells.
Further, it was noted that KD of IFITM1 had a small effect; however, silencing of IFITM3 resulted in 2- to 31-fold depletion of viral RNA production.
Interestingly, the production of infectious SARS-CoV-2 particles in Calu-3 cells reduced to near background levels upon silencing of IFITM2. Furthermore, the replication of SARS-CoV-2 VOC in iATII was reduced by the antibody targeting the N-terminus of IFITM2 in a dose-dependent manner.
"IFITMs (especially IFITM2) are also critical cofactors for efficient replication of current SARS-CoV-2 VOCs including the dominant Delta variant."
Conclusions
The study's findings suggest that endogenous IFITMs (especially IFITM2) are important cofactors for the effective replication of SARS-CoV-2 VOCs, including the dominant Delta variant. It was noted that the SARS-CoV-2 Delta variant replicated at a 30-times higher rate in comparison to the early NL-02-2020 isolate in iATII cells. Thus, the dependency of IFITM2 maintained by VOCs can be further explored as a target for therapeutic or preventive approaches.
Efficient inhibition of the SARS-CoV-2 Delta VOC by an a-IFITM2 antibody illustrates that IFITM2 may have a crucial role in SARS-CoV-2 transmission as well as pathogenesis.
"Our observation that IFITM2 dependency is maintained by VOCs also further underlines that this cellular "antiviral" factor represents an interesting target for therapeutic or preventive approaches."
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- IFITM dependency of SARS-CoV-2 variants of concern. Rayhane Nchioua, Annika Schundner, Dorota Kmiec, Caterina Prelli Bozzo, Fabian Zech, Lennart Koepke, Manfred Frick, Konstantin M. J. Sparrer, Frank Kirchhoff, bioRxiv, 2021. doi: https://doi.org/10.1101/2021.11.17.468942, https://www.biorxiv.org/content/10.1101/2021.11.17.468942v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Antibodies, Antibody, Cancer, Cell, Cell Line, Cell Sorting, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Fluorescence, Genes, Immunodeficiency, Influenza, Interferon, Lung Cancer, Pandemic, Propagation, Protein, Receptor, Remdesivir, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, siRNA, Syndrome, Virus, Western Blot

Written by
Saurabh Chaturvedi
Saurabh Chaturvedi is a freelance writer from Jaipur, India. He is a gold medalist in Masters in Pharmaceutical Chemistry and has extensive experience in medical writing. He is passionate about reading and enjoys watching sci-fi movies.
Source: Read Full Article