In a recent study posted to the bioRxiv* preprint server, scientists analyzed the necessity of revamping the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines to tackle the current dominant circulating viral variants.
 Study: Modeling suggests that multiple immunizations or infections will reveal the benefits of updating SARS-CoV-2 vaccines. Image Credit: klss / Shutterstock
Study: Modeling suggests that multiple immunizations or infections will reveal the benefits of updating SARS-CoV-2 vaccines. Image Credit: klss / Shutterstock
Background
The SARS-CoV-2 Omicron (OM) variant, which emerged at the end of 2021, is far more transmissible than the initial Wuhan (WU) strain. Omicron also contains numerous spike (S) protein mutations facilitating it to evade antibody responses to previous variants and existing Wuhan-based vaccines.
A study by Gagne et al. demonstrated that administering three doses of the vaccine centered on the SARS-CoV-2 ancestral Wuhan strain and two Wuhan isolate-based vaccine doses plus a booster dose with the updated vaccine based on the SARS-CoV-2 Omicron variant imparted comparable antibody titers against Omicron. It also showed that both vaccination regimens generated similarly high antibody levels against the Wuhan strain versus the Omicron variant. The research suggests a reformulation of the vaccine may not be necessary in order to deal with the Omicron strain. Besides, the timing of vaccine updates for evolving pathogens like SARS-CoV-2 is unknown.
About the study
In the present paper, the researchers employed computational simulations to increase the knowledge of the rules for enhancing CoV disease 2019 (COVID-19) vaccination responses to novel SARS-CoV-2 variants.
Computer models like the one used in this research could explain the complicated outcomes that result from interactions between various components. Integrating computational modeling to summarize trends observed in several datasets could thus play a significant role. Further, it should preferably be done iteratively, with the models being used to comprehend the available data and proposing experimental evaluations that could permit model rejection or refinement.
Gagne et al.'s findings and data from previous SARS-CoV-2 and influenza research were analyzed using computational models built by the team. They examined the significant aspects of increasing immunity after vaccination with the SARS-CoV-2 Omicron- and ancestral strain-based vaccines. In addition, the scientists analyzed the dynamics of B cells, antigens, and antibodies in the current simulations to figure out why both Omicron- and Wuhan strain-based vaccines had equal antibody concentrations to Omicron after the third shot. They then used this model to determine what would happen following more vaccinations/infections. Furthermore, the authors assessed how the model can be empirically evaluated.
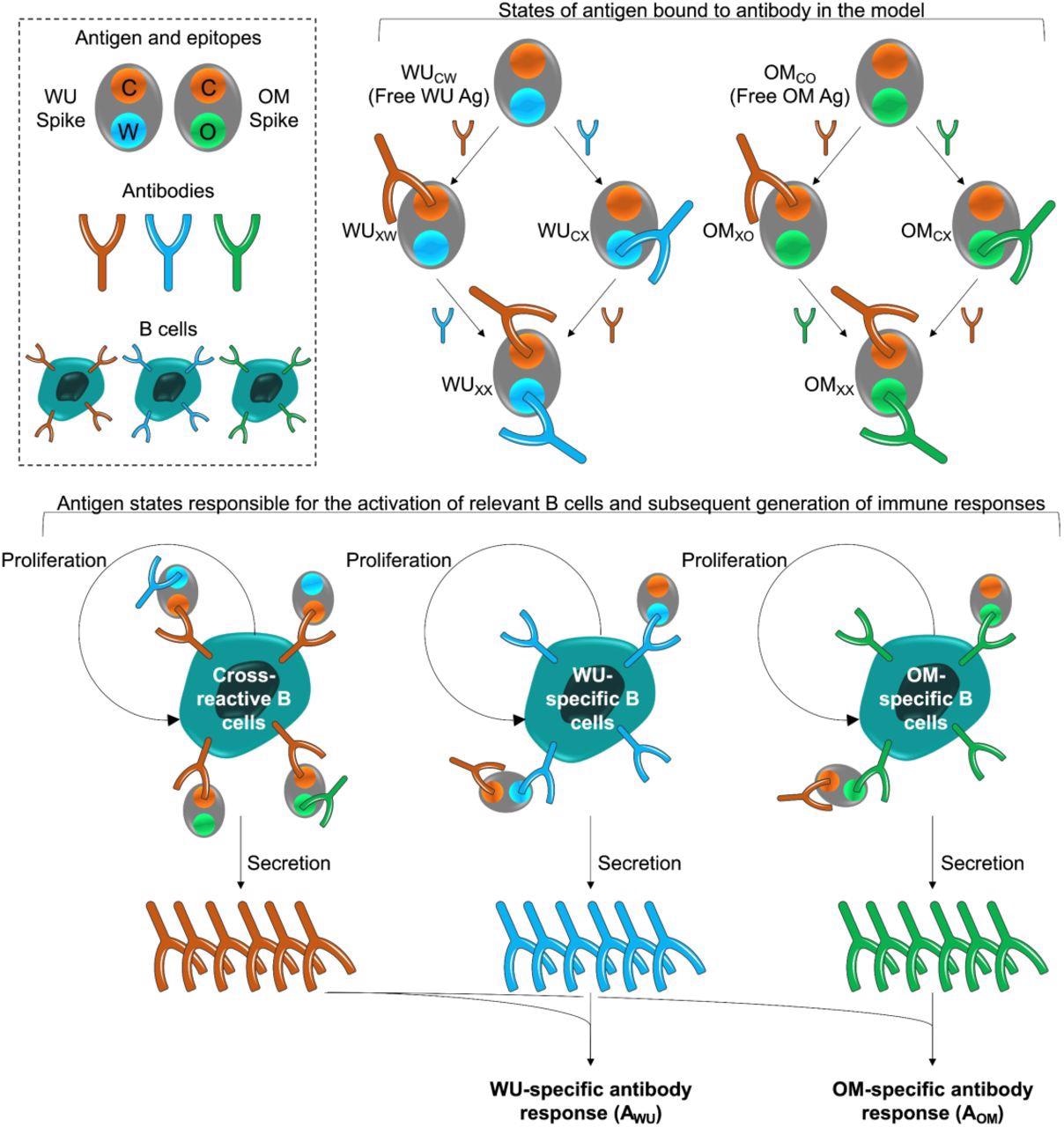
Model schematic. The box at the top left shows the epitopes of the WU- and OM-vaccines. Epitope C (shown in orange) is common to both vaccines. Epitopes W (blue) and V (green) are unique to the WU and OM respectively. Antibodies specific to these epitopes can bind to these epitopes and prevent them from stimulating B cells for the same epitope. The different antigen states generated and the B cells they stimulate are shown in the top right and bottom panels respectively. The bottom panel illustrates that binding of antigen to B cells stimulates their clonal expansion and the production of antibodies.
Results and discussions
The current mathematical model indicated that WU-WU-OM and WU-WU-WU vaccinations lead to similar antibody concentrations to Omicron since such a response was characterized by relatively strong secondary or recall responses to common epitopes shared by Omicron and Wuhan isolates. The intensity of this secondary reaction conceals the much lower primary response to novel Omicron epitopes that appear initially after third-time immunization with the Omicron vaccine yet not with the Wuhan strain-based vaccine.
Repeated boosts with an Omicron-based vaccination, such as the third, fourth, and fifth doses, resulted in substantially greater antibody levels for epitopes specific to Omicron, leading to a much greater total titer to Omicron. The current simulations projected that repeated immunizations with the revised COVID-19 vaccine would be required to improve responses to the novel epitopes found in the antigens of new viral variants.
This model recommended a vital experiment to enable rejection or validation of the paradigm, involving giving the primates in Gagne et al.'s study one extra vaccination shot (fourth) with Omicron. The model projected that the animals that received WU-WU-OM-OM vaccines would have much greater Omicron antibody titers than WU-WU-WU-WU. Additionally, after WU-WU-OM vaccination, the scientists expected considerably greater antibody titers to the Omicron variant after spontaneous infection with Omicron.
In light of the present findings, the researchers hypothesized that most antibodies produced after the first shot of a vaccine upgraded to match a new viral mutant were specific to the common antigens. Moreover, only after a second vaccination with the same vaccine elevated antibody responses specific for epitopes peculiar to the novel variant emerge.
Besides, updating the vaccination to match novel viral variants may improve individual antibody responses facilitating better targeting of future variants that evolve from the current Omicron strain.
Finally, the researchers suggested that the at-risk populations would benefit from receiving two doses of updated vaccinations, not just for COVID-19 but also for influenza.
Conclusions
Altogether, the computational models utilized in this work focused on whether the COVID-19 vaccines should be upgraded to the current SARS-CoV-2 variant, specifically Omicron.
The current models confirm previous findings, but they also imply that immunization with the revised COVID-19 vaccine results in qualitatively altered humoral immunity, i.e., a tiny portion of it being specific for novel viral variant epitopes. According to current simulations, these novel variant-specific responses might dominate after successive vaccination or infection with either the present or upcoming viral variants.
Additionally, the researchers proposed crucial studies to either reject or confirm the model. They recommended two doses of the updated vaccination approach to assess if the vaccine needs to be updated and vaccinate at-risk people.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Modeling suggests that multiple immunizations or infections will reveal the benefits of updating SARS-CoV-2 vaccines; Rajat Desikan, Susanne L Linderman, Carl W. Davis, Veronika I Zarnitsyna, Hasan R Ahmed, Rustom Antia. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.05.21.492928, https://www.biorxiv.org/content/10.1101/2022.05.21.492928v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Antibodies, Antibody, Antigen, Computational Modeling, Coronavirus, Coronavirus Disease COVID-19, covid-19, immunity, Immunization, Influenza, Omicron, Protein, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Vaccine

Written by
Shanet Susan Alex
Shanet Susan Alex, a medical writer, based in Kerala, India, is a Doctor of Pharmacy graduate from Kerala University of Health Sciences. Her academic background is in clinical pharmacy and research, and she is passionate about medical writing. Shanet has published papers in the International Journal of Medical Science and Current Research (IJMSCR), the International Journal of Pharmacy (IJP), and the International Journal of Medical Science and Applied Research (IJMSAR). Apart from work, she enjoys listening to music and watching movies.
Source: Read Full Article