DNA replication is required to maintain the integrity of genomic information. This article describes the process of DNA replication, in a step-by-step manner.
 Soleil Nordic | Shutterstock
Soleil Nordic | Shutterstock
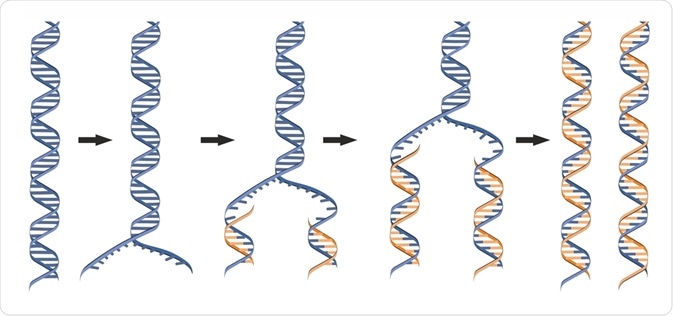
DNA replication requires unwinding of the complementary two-stranded structure of DNA. This process is mediated by the severance of the hydrogen bonds that hold the bases together; the result is the formation of two single strands.
The resultant Y-shaped appearance in this region of DNA is called the replication fork. The initiation of replication occurs at specific sites called the origin of replication (ori). Following the establishment of the replication fork is the replisome, several factors which enable replication to take place.
DNA Polymerases are one such crucial factor. They are multi-subunit enzymes that participate in the process of DNA replication in the cell. They catalyze the addition of nucleotides onto existing DNA strands. There are many families of DNA polymerase that play a role in DNA replication; there are at least 15 in humans and are required at different points during the process.
Polymerase function during DNA replication
DNA polymerase enzymes typically work in a pairwise fashion; each enzyme replicates one of the two strands that comprise the DNA double helix. These are called the leading strand and lagging strand and are named according to the relative speed at which they are replicated.
The replicated strands are synthesized using the leading and lagging strands as templates. Consequently, the two new double-stranded DNA molecules produced consist of one strand from the original helix (either the leading or lagging strand) and one new strand. This process is called semi-conservative replication and is essential as it permits genetic information to be transmitted from generation to generation.
The activities of both DNA polymerases are coordinated by two structures called the sliding clamp loader and the sliding clamp. The sliding clamp loader contacts single-stranded binding proteins that coat the separated helix as well as the sliding clamp.
Two sliding clamps encircle the two strands of DNA, and together with accessory proteins called the clamp loader complex, provide a stable binding site for the two DNA polymerases. The unwound single-stranded DNA templates move toward the complex; the behavior of the clamp loader on the leading and lagging strand differ because of a property called directionality.
This is determined by the orientation of the phosphate bond and characterized by the conventions 5’ to 3’ and 3’ to 5’. Each of the two strands of the helix necessarily possess opposite directionality; this is essential for base pairing to occur. Their pairing is also referred to as antiparallel.
DNA polymerase synthesizes only in a 5′ to 3′ direction. Consequently, the strand with the complementary 3’ to 5’ directionality, the leading strand, is synthesized as one continuous piece. Conversely, the strand with 5’ to 3’ directionality is synthesized as a series of small fragments called Okazaki fragments.
The lagging strand orientation of 5’ to 3’ is incompatible with DNA polymerase; to accommodate this requirement, the clamp loader must continually release and reattach at a new location. This requires the lagging strand to bubble out from the replisome.
Polymerases for DNA repair
Several polymerases exist in both prokaryotes and eukaryotes. They provide polymerase activity under two broad categories; normal replication and repair. Under conditions of normal replication, DNA polymerase corrects errors by 3′ → 5′ exonuclease activity.
Outside of normal replicative events, DNA repair is an ongoing process that is necessary to maintain the integrity of the genome. Both endogenous and exogenous insults result in damaged DNA; for example, single-strand and double-strand breaks, strand crosslinking, base loss and base modification.
Multiple pathways exist to repair these DNA damage events in a selective manner. These include mismatch repair, nucleotide excision repair, base excision repair, double-strand break repair and inter-strand cross-link repair. The biochemical difference that exists between these polymerases allows them to fulfill distinct roles under these specific conditions of repair.
Sources
- http://www-lehre.img.bio.uni-goettingen.de/Vorlesung_SS_2007/
- http://dnareplication.cshl.edu/content/free/chapters/15_wang.pdf
- www.informatics.indiana.edu/…/lecture_notes_19b.pdf
- https://www.sciencedirect.com/science/article/pii/S006532330469005X
- https://www.sciencedirect.com/science/article/pii/B9780123850157000041
Further Reading
- All DNA Replication Content
- DNA Replication and Repair
- Homologous Recombination
- Mechanisms of DNA Repair
- Mechanism of DNA Synthesis
Last Updated: Apr 11, 2019

Written by
Dr. Ananya Mandal
Dr. Ananya Mandal is a doctor by profession, lecturer by vocation and a medical writer by passion. She specialized in Clinical Pharmacology after her bachelor's (MBBS). For her, health communication is not just writing complicated reviews for professionals but making medical knowledge understandable and available to the general public as well.
Source: Read Full Article