In an article published in Scientific Reports, researchers affiliated with the University of São Paulo (USP) in Brazil describe experiments performed with mice that show how genetic factors can determine whether an individual is susceptible or resistant to leishmaniasis. According to the authors, the results will contribute to a better understanding of leishmaniasis in humans and help to clarify why only some of those individuals infected develop the disease.
Leishmaniasis is caused by protozoan parasites of the genus Leishmania. Depending on the species involved, infection leads to skin ulcerations (cutaneous leishmaniasis) or lesions in internal organs such as the liver and spleen (visceral leishmaniasis). The parasites are transmitted to humans and other mammals by insect bites. There are no available vaccines for the disease, and its treatment is lengthy, expensive and complicated.
“We set out to observe gene regulation in our study model in order to investigate the strategy used by an organism capable of resisting infection and find out how it differs from a susceptible organism in this regard,” said Lucile Maria Floeter-Winter principal investigator of the project in the Physiology Laboratory of the University of São Paulo’s Bioscience Institute (IB-USP).
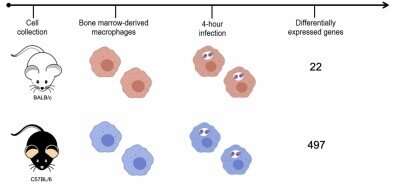
The researchers used two mouse strains: BALB/c, which is naturally sensitive to the parasite, and C57BL/6, which is naturally resistant. They infected both strains with Leishmania amazonensis, the species that causes cutaneous leishmaniasis. In the article, they stress that the identification of molecular markers for resistance to the disease can be useful for diagnosis, prognosis and clinical intervention.
Gene expression mapping
Leishmania species have developed the strategy of infecting host macrophages, which are one of the types of immune cells that should be involved in combatting the parasite. Infected macrophages may burst, releasing the multiplying protozoans, which then infect other nearby macrophages. At this point, the infection is considered established. Alternatively, the parasites are killed by the macrophages, and the disease fails to progress.
To create an experimental model of infection by the parasite, the researchers took cells from the bone marrow of the mice, differentiated these cells into macrophages, and infected them. After four hours, the RNA of the infected macrophages was extracted and sequenced. Gene expression mapping based on the analysis of the transcriptomes of both murine strains showed which genes were expressed during the first four hours of infection in each group.
“We focused on the start of the infection, when the response mechanisms are being triggered,” said Juliana Ide Aoki, a researcher at IB-USP and lead author of the article, supported by a postdoctoral scholarship)from FAPESP.
“When the organism is infected, it alerts the macrophages to produce a group of molecules that are responsible for responding to the infection. Our analysis of the transcriptome showed which segments of the genome understood this signal and effected a response to combat the parasite,” Floeter-Winter said.
A total of 12,641 genes were expressed by macrophages cultured in the laboratory, but only 22 genes were found to be expressed in response to the parasite in the susceptible murine strain (BALB/c), compared with 497 in the resistant strain (C57BL/6).
“This is the key finding from our study. The susceptible organism activated only a few genes and was unable to stop the infection. The resistant organism activated a large number of genes, inducing the production of molecules to control the infection, and succeeded in doing so,” Floeter-Winter said
“Our results show that the development of the disease depends on the genetics of the host and not just of the parasite. This may explain why the infection progresses in some patients whereas others are resistant.”
According to Floeter-Winter, the mice received the same treatment and food, ruling out the influence of environmental factors on the results. Future research may be aimed at determining why the resistant strain (C57BL/6) activates more genes to combat the infection.
The identification of molecules present in an infected organism that is capable of controlling the infection will help scientists to suggest markers for the evaluation and determination of the prognosis of human patients. “For example, it would be possible to see which molecules an infected patient expresses and to predict whether the infection will last a relatively long time, whether it will be severe, and whether the patient is producing molecules that combat it,” she said.
Furthermore, the study’s scientific contribution can be extrapolated to other aspects of the disease. “A better understanding of how Leishmania establishes infection extends our knowledge of response mechanisms, which can be used in treating other infectious diseases and can help other researchers conduct studies with different models,” Floeter-Winter said.
Source: Read Full Article