During the early phases of the coronavirus disease 2019 (COVID-19) pandemic, the speed and approach of different countries' responses to the pandemic were essential in keeping case numbers low.
Generally, governments that were slow to impose restrictions or prevent inward travel struggled with rising hospitalizations early on. At the same time, countries such as Singapore and New Zealand were widely applauded for their effective methods. In several countries, these preventative measures included mandatory isolation for those traveling into the country, either to allow time for the individual to be tested or time for symptoms to show.

Study: Early detection of SARS-CoV-2 variants using traveler-based genomic surveillance at four US airports. Image Credit: Thanakorn.P / Shutterstock.com
In a recent study published on the preprint server medRxiv*, researchers from Ginkgo Bioworks discuss their efforts to detect the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants in travelers entering the United States.
About the study
Travelers arriving at either John F. Kennedy in New York or San Francisco airports on seven direct flights from India were monitored for approximately two months between September 29, 2021, and November 27, 2021. In addition, Hartsfield-Jackson Atlanta International airport was added between November 28, 2021, to January 23, 2022, wherein participation was offered to travelers arriving from South Africa, Nigeria, the United Kingdom, France, Germany, and Brazil, with about 50 flights arriving in Atlanta each day from these nations.
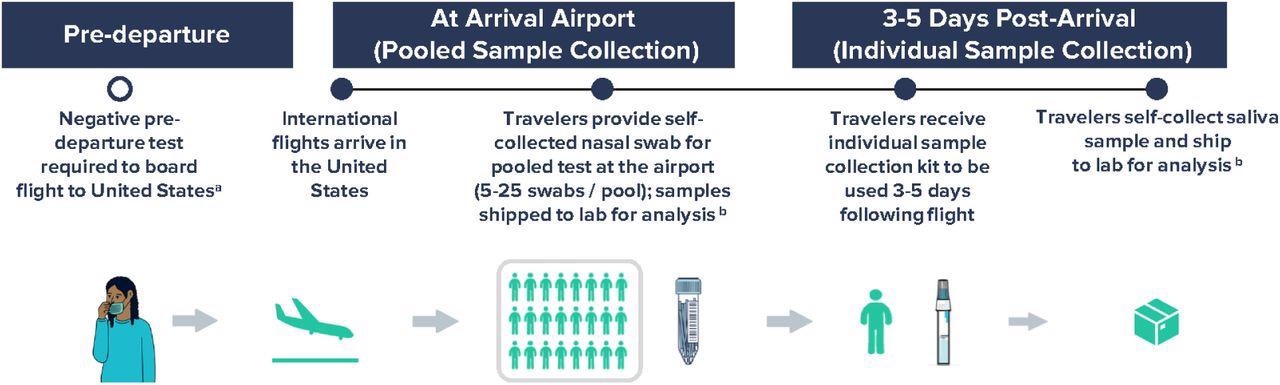
Workflow for airport pooled and individual sample collection to detect SARS-CoV-2 variants of concern as part of an international arriving traveler surveillance program, United States, September 29 – January 23, 2022. a Negative COVID-19 test was required to board a flight to the United States per CDC Order; the time window for pre-departure testing was shortened from 3 days to 1 day on December 6, 2021. b Analyses included RT-PCR testing of all samples within 24-48 hours of sample collection and whole-genome sequencing of all positive samples in a median of 11 days.
Participants were only eligible for inclusion if they were 18 years old or above and provided informed consent as well as answered demographic, clinical, and travel history questions.
Study participants were then asked to provide saliva and nasal samples three to five days after arrival. These samples then underwent reverse transcription-polymerase chain reaction (RT-PCR) testing for SARS-CoV-2, with all samples that tested positive then used for whole-genome sequencing and variant lineage determination. Fisher's exact tests assessed differences in pooled positivity rates across the two different collection periods and by country of origin.
Study findings
Taken together, about 16,000 travelers were enrolled in the current study, of about 161,000 of whom were eligible. A total of 1,454 samples pools were recovered, 221 of which tested positive for SARS-CoV-2.
SARS-CoV-2 positivity increased over time, from 1.8% during the September-November period to 20.9% after November 27, 2021. The positivity percentage of pooled samples was significantly different based on the country of origin, with flights from South Africa averaging 43.5%, while flights from India averaged 9.3% positivity.
The SARS-CoV-2 variants that were detected also varied over time, with all lineages detected before November 28, 2021, diverging from the Delta variant. other than one undetermined lineage. Comparatively, between November 28 and January 2022, only 5% of samples were Delta, whereas 27% were undetermined due to low sample sequencing coverage, and all remaining samples were Omicron.
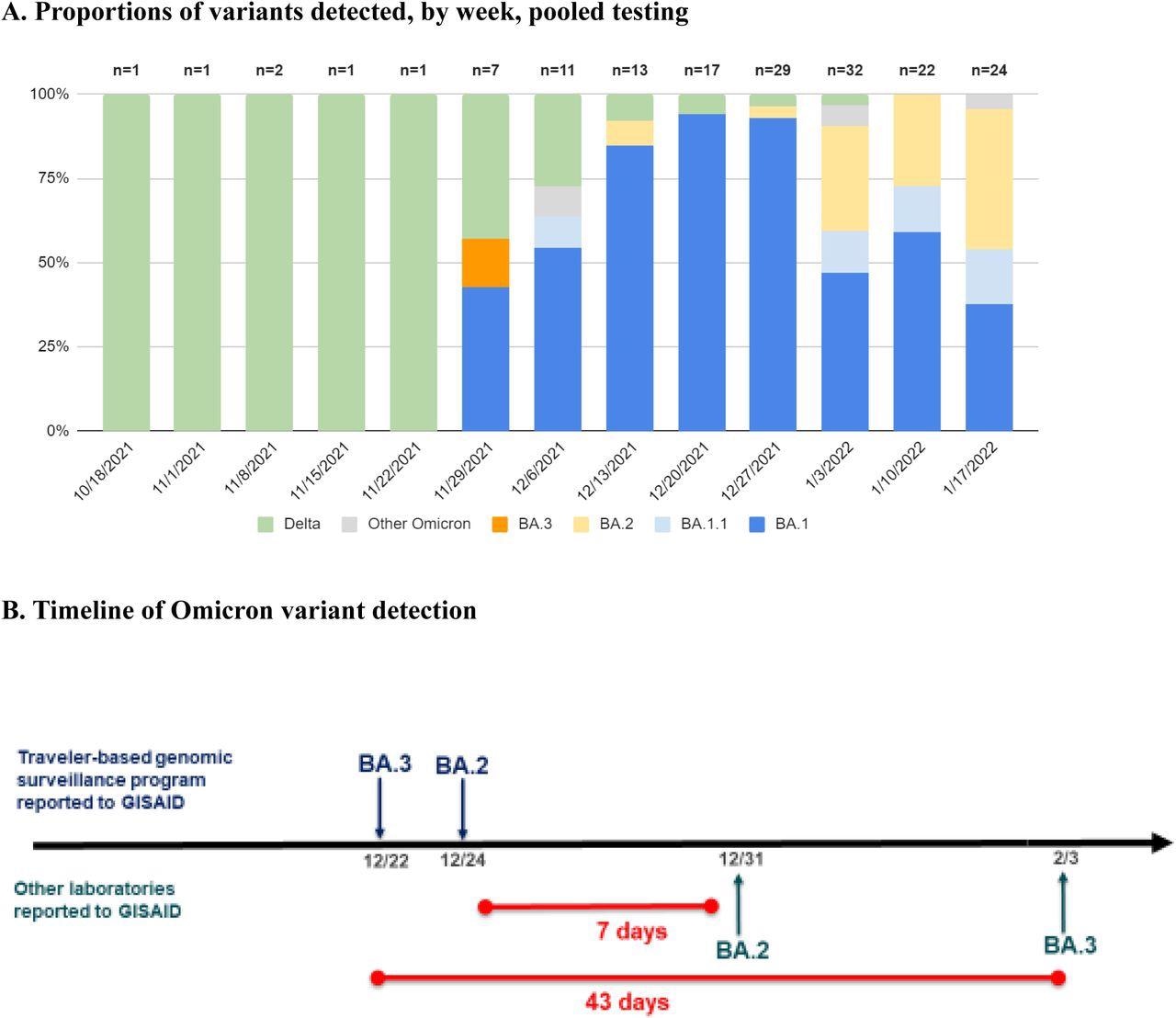
Proportions of variants detected among arriving international travelers and rapid reporting of Omicron subvariants. A, Identified variant lineage and sublineage proportions among sequenced samples by collection week, November 28, 2021 to January 23, 2022. B, Timeline of reported Omicron subvariants in the United States, comparing first reporting in GISAID of BA.2 and BA.3 by the traveler-based genomic surveillance program (in blue) compared to first reporting from other laboratories (in green). The program’s timeliness of first reporting of BA.2 (on December 24) and BA.3 (on December 22) was 7 and 43 days earlier, respectively, than next reporting in GISAID.
Out of the 145 identified Omicron sequences, 112 showed complete or partial spike gene target failure (SGTF); however, this was not observed in the BA.2 sublineages. The vast majority of Omicron sublineages detected were BA.1; however, BA1.1, BA.2, BA.3, BA.2 +Orf1a:M85, and BA.2 + S:R346K were all detected, as well as four Omicron samples where the sublineages of which could not be determined. One of the BA.2 samples recovered on December 14, 2021, was the first reported case of BA.2 in the United States, as was one of the BA.3 samples.
The program successfully identified new SARS-CoV-2 variants when they were imported into the U.S, including two Omicron sub-lineages that were detected before being reported elsewhere in the country. As expected, the number of positive tests increased significantly following Omicron emergence.
Interestingly, the number of positive tests also increased after December 6, 2021, when passengers were required to have a negative sample collected before departure. This could be due to poor sensitivity during testing or passengers arriving on particularly long-haul flights who became infective during travel. It is also possible that certain travelers presented fraudulent test results in order to fly.
Conclusions
The authors highlight the scalability and adaptability of their screening program and recommend that a similar method be adopted in the future. This would allow more rapid detection of new SARS-CoV-2 variants, as currently, many are only identified after they have already spread widely.
Additionally, the study approach allows for early warning systems in the event that a large number of positive cases from particular countries emerge. This would subsequently allow travel restrictions to be put in place when necessary, without having to rely on the sometimes inaccurate case numbers provided by certain governments.
Several Southeast Asian countries were lauded for low case numbers early on in the pandemic, despite their proximity to the country of origin. In fact, many of these nations credited the track and trace programs that were launched very early in the pandemic to their success. Therefore, more widespread adoption of this approach could help keep case numbers lower in future pandemics.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Wegrzyn, R. D., Appiah, G. D., Morfino, R., et al. (2022). Early detection of SARS-CoV-2 variants using traveler-based genomic surveillance at four US airports. medRxiv. doi:10.1101/2022.03.21.22272490. https://www.medrxiv.org/content/10.1101/2022.03.21.22272490v1.
Posted in: Device / Technology News | Medical Science News | Medical Research News | Disease/Infection News
Tags: Coronavirus, Coronavirus Disease COVID-19, covid-19, Gene, Genome, Genomic, Ginkgo, Omicron, Pandemic, Polymerase, Polymerase Chain Reaction, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription

Written by
Sam Hancock
Sam completed his MSci in Genetics at the University of Nottingham in 2019, fuelled initially by an interest in genetic ageing. As part of his degree, he also investigated the role of rnh genes in originless replication in archaea.
Source: Read Full Article