In a recent study published on the preprint server medRxiv*, researchers found that infection with one Omicron sub-lineage of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appears to induce strong protection against reinfection with the other Omicron sub-lineage for at least several weeks after the initial infection.
Study: Protection of Omicron sub-lineage infection against reinfection with another Omicron sub-lineage. Image Credit: i-am-helen / Shutterstock.com
SARS-CoV-2 infection with earlier variants provide poor protection against Omicron variant
Because antibodies raised by primary SARS-CoV-2 vaccination are waning in vaccinated populations, several reports of reinfections with SARS-CoV-2 variants have emerged around the world, thereby challenging the global response to the pandemic. The latest variant Omicron and its two major sub-lineages, BA.1 and BA.2, harbor multiple mutations in the spike protein that allow immune escape. As a result, the SARS-CoV-2 Omicron variant is responsible for a majority of reinfections occurring across the globe.
While SARS-CoV-2 infection with earlier variants has provided high levels exceeding 85% protection against reinfections with the Alpha, Beta, and Delta variants, protection elicited against reinfection with the Omicron BA.1 sub-lineage is inferior at less than 60%.
Currently, Qatar is experiencing a significant wave of new coronavirus disease 2019 (COVID-19) cases that were previously due to the BA.1 sub-lineage of Omicron variant, and are now dominated by the BA.2 sub-lineage. Thus, the researchers in the current study investigated the protective efficacy of infection with one sub-lineage against reinfection with the other sub-lineage between December 19, 2021, to February 21, 2022, in Qatar.
About the study
Based on information from Qatar federated databases for COVID-19, two retrospective cohort studies were conducted. Whereas one study assessed the effectiveness of BA.1 infection against reinfection with BA.2 sub-lineage and included 20,197 individuals, the other study assessed the effectiveness of BA.2 infection against reinfection with BA.1 and included 100,925.
An S-gene “target failure” (SGTF) case using the reverse-transcriptase qualitative polymerase chain reaction (RT-qPCR) assay was considered as a BA.1 sub-lineage infection. Conversely, non-SGTF cases were considered as the BA.2 sub-lineage infection.
Since the Omicron wave started recently, the team did not consider the already established definition of ‘SARS-CoV-2 reinfection,’ which is a documented infection more than 90 days after an earlier infection. Rather, an analysis of durations of follow-up was conducted to identify the longest time interval possible between reinfections, while maintaining the precision of estimates. Reinfection was thus redefined as reporting of infection more than 35 days after the prior infection.
The study population was exact-matched by sex, age group, and nationality to the uninfected controls.
The cumulative incidence rate of infection in each cohort, hazard ratio comparing the incidence in case vs control cohorts, and effectiveness of protection were calculated and statistically analyzed.
Study findings
In the BA.1-against-BA.2 study, the cumulative incidence of infection was estimated to be 0.03% for the BA.1-infected cohort and 0.62% for the uninfected-control cohort 15 days after the start of follow-up. The adjusted hazard ratio for infection was estimated at 0.05, whereas the effectiveness of BA.1 infection against reinfection with BA.2 was estimated to be 94.9%.
In the BA.2-against-BA.1 study, the cumulative incidence of infection was estimated to be 0.03% for the BA.2-infected cohort and 0.17% for the uninfected-control cohort 15 days after the start of follow-up. The adjusted hazard ratio for infection was estimated at 0.14, with the effectiveness of BA.2 infection against reinfection with BA.1 estimated to be 85.6%.
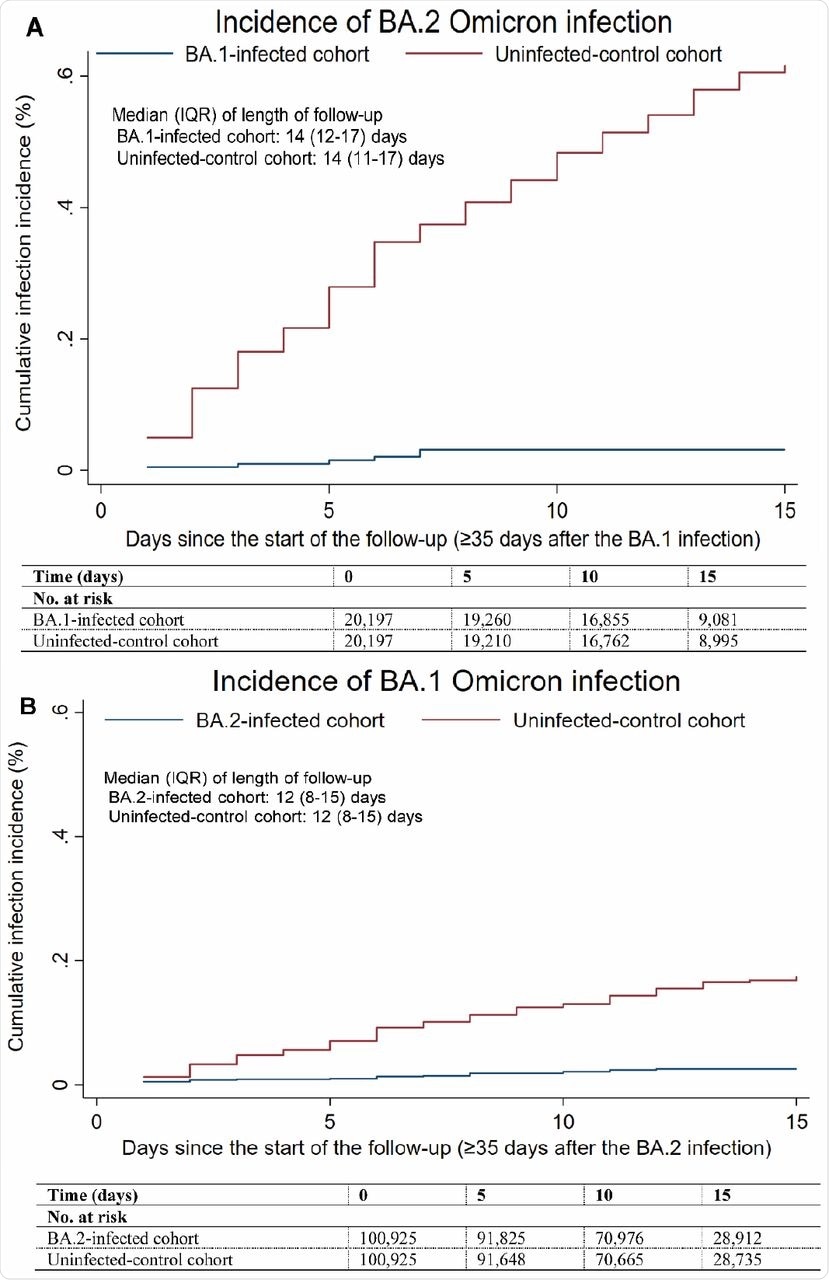 Cumulative incidence of A) BA.2 and B) BA.1 Omicron infections in the BA.1-against-BA.2 and BA.2-against-BA.1 studies, respectively.
Cumulative incidence of A) BA.2 and B) BA.1 Omicron infections in the BA.1-against-BA.2 and BA.2-against-BA.1 studies, respectively.
Conclusions
Regardless of the sub-lineage of the infecting or reinfecting Omicron variant, a strong but not full protection was imparted against reinfection.
It is remarkable that incidence of reinfection, regardless of sub-lineage, was much lower in the BA.1-infected and BA.2-infected cohorts than incidence of infection in the corresponding uninfected-control cohorts.”
Based on the findings, the team suggests that the natural immunity of SARS-CoV-2 variants be clustered into two groups including an early non-Omicron variant group and recent Omicron BA.1 and BA.2 sub-lineages group. Within each group, strong protection is imparted against reinfection, with effectiveness exceeding 85%. However, between the groups, the protection might not exceed 60%, as per the recent reports.
The protection efficacy of Omicron infection with either variant exceeds 85%, which is similar to the protection observed for infection with the wild-type or early variants of SARS-CoV-2 such as the Alpha, Beta, and Delta variants against reinfection with these strains of SARS-CoV-2.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Chemaitelly H., Ayoub, H. H., Coyle, P., et al. (2022). Protection of Omicron sub-lineage infection against reinfection with another Omicron sub-lineage. medRxiv. doi:10.1101/2022.02.24.22271440. https://www.medrxiv.org/content/10.1101/2022.02.24.22271440v1.
Posted in: Medical Science News | Medical Research News | Medical Condition News | Disease/Infection News
Tags: Antibodies, Assay, Coronavirus, Coronavirus Disease COVID-19, covid-19, Efficacy, Gene, immunity, Omicron, Pandemic, Polymerase, Polymerase Chain Reaction, Protein, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome

Written by
Namita Mitra
After earning a bachelor’s degree in Veterinary Sciences and Animal Health (BVSc) in 2013, Namita went on to pursue a Master of Veterinary Microbiology from GADVASU, India. Her Master’s research on the molecular and histopathological diagnosis of avian oncogenic viruses in poultry brought her two national awards. In 2013, she was conferred a doctoral degree in Animal Biotechnology that concluded with her research findings on expression profiling of apoptosis-associated genes in canine mammary tumors. Right after her graduation, Namita worked as Assistant Professor of Animal Biotechnology and taught the courses of Animal Cell Culture, Animal Genetic Engineering, and Molecular Immunology.
Source: Read Full Article