Final results from a study of a blood test that can detect more than 50 types of cancer have shown that it is accurate enough to be rolled out as a multi-cancer screening test among people at higher risk of the disease, including patients aged 50 years or older, without symptoms.
In a paper published in the leading cancer journal Annals of Oncology today, researchers report that the test accurately detected cancer, often before any symptoms arose, while having a very low false positive rate. The test also predicted where in the body the cancer is located with a high degree of accuracy, which could help doctors choose effective diagnostic tests.
GRAIL, Inc. (California, U.S.), the company developing and funding the research, has now made the multi-cancer early detection test available in the U.S. by prescription only, and to complement other, existing screening methods such as those for breast, cervical, prostate, lung and bowel cancers. Many of the cancers that the test is capable of detecting do not have screening tests available, such as liver, pancreatic and oesophageal cancers, which are among the most deadly and where early detection could make a real difference.
First author of the paper, Dr. Eric Klein, chairman of the Glickman Urological and Kidney Institute, Cleveland Clinic, Cleveland, U.S., said: “Finding cancer early, when treatment is more likely to be successful, is one of the most significant opportunities we have to reduce the burden of cancer. These data suggest that, if used alongside existing screening tests, the multi-cancer detection test could have a profound impact on how cancer is detected and, ultimately, on public health.”
The test involves taking a sample of blood from each patient and analysing it for DNA, known as cell-free DNA (cfDNA), which tumours (and other cells) shed into the blood. Genomic sequencing is used to detect chemical changes to the DNA called “methylation” that control gene expression, and a classifier developed with machine learning (artificial intelligence) uses these results to detect abnormal methylation patterns that suggest cancer is present. In addition, the machine learning classifier can predict where in the body the cancer is located. Results are available within ten business days from the time the sample reaches the lab.
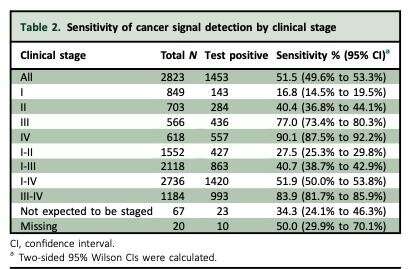
The third and final sub-study of the Circulating Cell-free Genome Atlas (CCGA) study reported today investigated the performance of the test in 2,823 people already diagnosed with cancer and 1,254 people without cancer. It detected cancer signals from more than 50 different types of cancer and found that across all four cancer stages (I, II, III, IV), the test correctly identified when cancer was present (the sensitivity or true positive rate) in 51.5% of cases. The test’s specificity (the true negative rate) was 99.5%, meaning that the test wrongly detected cancer (the false positive rate) in only 0.5% of cases.
Sensitivity of the test was 67.6% overall across stages I-III in 12 pre-specified cancers that account for two-thirds of cancer deaths in the U.S. each year (anal, bladder, bowel, oesophageal, stomach, head and neck, liver and bile duct, lung, ovarian and pancreatic cancers, lymphoma and cancers of white blood cells such as multiple myeloma), and it was 40.7% overall in more than 50 cancers.
For all cancers, detection improved with each cancer stage with a sensitivity rate of 16.8% at the early stage I, 40.4% at stage II, 77% at stage III and 90.1% at stage IV—the most advanced stage when symptoms are often showing.
The sensitivity varied by type of cancer. In solid tumours that do not have any screening options, such as oesophageal, liver and pancreatic cancers, overall sensitivity of the test was twice that for solid tumours that do have screening options, such as breast, bowel, cervical and prostate cancers: 65.6% compared to 33.7%. Overall sensitivity in cancers of the blood, such as lymphoma and myeloma, was 55.1%.
In addition, the multi-cancer early detection test correctly identified the tissue in which the cancer was located in the body in 88.7% of cases.
Dr. Klein said: “We believe that cancers that shed more cfDNA into the bloodstream are detected more easily. These cancers are also more likely to be lethal, and prior research shows that this multi-cancer early detection test more strongly detects these cancer types. Cancers such as prostate shed less DNA than other tumours, which is why existing screening tests are still important for these cancers.”
In order to understand how the test would perform when used to screen populations, the researchers estimated its positive predictive value (PPV) – the proportion of cases correctly identified as cancer among those with a positive result—as well as the negative predictive value (NPV) – those correctly identified as not having cancer. The PPV was 44.4% among people most likely to develop cancer, those aged 50-79, and the NPV was 99.4%.
Dr. Klein concluded: “These data add to a growing body of literature that supports the use of next-generation sequencing for the detection of cell-free DNA in blood samples as a tool for earlier detection of common cancers that account for a significant number of deaths and other health problems worldwide. In addition, a screening test that requires only a simple blood draw could provide an option for communities that have poor access to medical facilities. I’m excited about the potential impact this approach will have on public health.”
Researchers are continuing to collect additional data from the test in large, prospective studies in the U.S. (STRIVE, PATHFINDER and REFLECTION studies) and the UK (SUMMIT study), and to examine its feasibility for screening populations. GRAIL has also established a partnership with the UK’s National Health Service to investigate the multi-cancer early detection test’s clinical and economic performance in approximately 165,000 eligible patients, starting later this year.
A strength of the CCGA study is that, overall, it includes a total of 15,254 participants from 142 clinics in North America, helping to ensure the results can be generalisable to a diverse population. The participants in this final sub-study had not been included in the earlier, development stages of the test to ensure accurate estimations of performance. Limitations of this sub-study include: if blood samples were collected from cancer patients after they had had a biopsy, this could increase the proportion of cfDNA in the blood compared to before the biopsy; CCGA is a case control study and may not fully reflect how the test would perform in population screening conditions (this is being evaluated in the PATHFINDER study); and some inaccuracy occurred in the detection of the tissue of cancer origin for cancers that are driven by the human papilloma virus (HPV), such as cancers of the cervix, anus, and head and neck.
Source: Read Full Article