Many of the diseases caused by point mutations have no established therapeutic approaches. Prof. Tsukahara and colleagues (Japan Advanced Institute of Science and Technology) are studying a therapeutic method using artificial RNA editing. Artificial site-directed RNA editing is an important technique for modifying genes and ultimately regulating protein function. We are trying to modify the genetic code of transcripts (RNA) by artificial RNA editing for the treatment of genetic disorders.
Although genome editing is drawing attention as a gene repair technology, genome editing such as CRISPR/Cas9 may result in permanent alterations, potentially affecting multiple loci. Currently, it is very difficult to perform accurate genome editing in all targeted cells in vivo. Although it is possible to edit the genome in a fertilized egg, embryo, or cells, it is not suitable for gene therapy in humans. Moreover, genome editing raises ethical concerns. Therefore, we believe that genome editing is a suitable method for “ex vivo” techniques, or for use in fertilized eggs, but not throughout a patient’s body. In contrary, changes resulting from the RNA editing are not permanent because they do not affect the genome sequence, and can be context-specific. Therefore, for the purpose of therapeutic treatments, RNA editing is preferable to genome editing, says Prof. Tsukahara.
RNA editing is a physiological process and widespread in living organisms to produce various proteins with different functions from a single gene. In mammals, C or A of the RNA chain is base sequence-specifically hydrolytically deaminating, whereby C is replaced by U and A by I (inosine). Since I forms a Watson-Crick base pair with C, the genetic code is synonymous with G. These base conversions occur as a result of deamination of A or C, which has been found to be catalyzed by ADAR and APOBEC family enzymes. Recently, various techniques for RNA restoration by artificial RNA editing using ADARs have been reported.
In this paper, we review recent findings regarding the application of ADARs to restoring the genetic code along with different approaches involved in the process of artificial RNA editing by ADAR. We have also addressed comparative studies of the various isoforms of ADARs. Therefore, we will try to provide a detailed overview of the artificial RNA editing and the role of ADAR with a focus on the enzymatic site directed A-to-I editing.
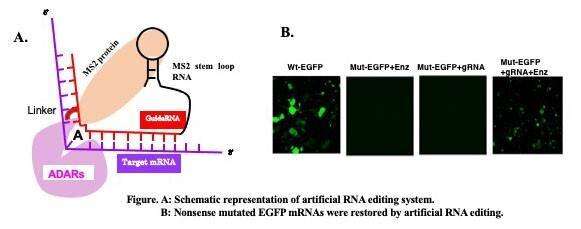
Most of artificial RNA editing systems use an active site of catalytic enzymes, ADARs, and a guide RNA complementary to the target to recruit an active site to the target RNA. One approach to artificial RNA editing is the use of chemical methods. Vogel and colleagues employed SNAP tag to join ADARs with a guide RNA and reported that the system is functional in vitro and in vivo. However, this technique requires a continuous supply of effector molecules to be effective. It is also known to use RNA-binding proteins to bind guide RNAs to enzymes. Two types of tethering system originating from bacteriophage are typically used in eukaryotes: the Lambda N system and MS2 system. Use of the ADAR enzyme with the MS2 system enables restoration of the genetic code, and holds promise for gene therapy.
Source: Read Full Article