Many big pharma companies were able to roll out vaccines against SARS-CoV-2 in record time by retooling their existing experimental mRNA gene therapies to carry a new payload. Whereas these cancer vaccines originally coded a single RNA molecule neoantigen concatemer that could be optimally presented on specific HLA receptors to enhance CD8+ T cell anti-tumor responses, they can now deliver optimized versions of the spike protein receptor binding domain to arouse production of protective neutralizing antibodies.
But things have not ended here. These same companies have also been hard at work repurposing their radically streamlined drug discovery pipelines to screen small molecule inhibitors that might block SARS infection, life cycle, or inflammatory sequelae in record time. As the winners of the vaccine battles and the treatment challenges now begin to emerge, an inspired industry now reignites the war on cancer with renewed vigor and purpose.
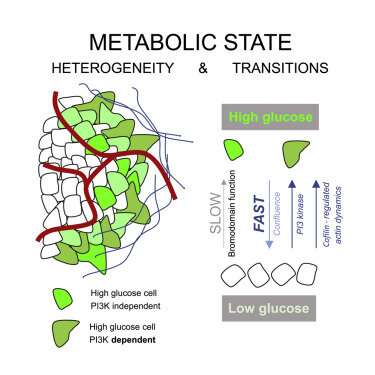
Monday, we reported on new technologies, like Mission Bio’s Tapestri Platform, that make it possible to simultaneously evaluate protein biomarkers and genetic defects in single cells throughout the entire lifecycle of the cancer. While this kind of information is critical for adjusting therapies as new resistant clones arise within heterogeneous tumors, there is more to the story. A cell’s genetic profile indicates what it is capable of doing, i.e., what substrates it could potentially use, and what substrates they absolutely need—but not so much about what the cells are actually doing at any given time. In other words, it indicates how fast they are metabolizing substrates—at the topmost level, this would be either via glycolysis, aerobic glycolysis or mitochondrial oxidative phosphorylation.
Imaging modalities such as positron emission tomography (18F-FDG PET), computed tomography, and hyper-polarized magnetic resonance imaging (MRI) have been invaluable tools for distinguishing between metabolic states in tumors, but they completely lack sufficient resolution to study heterogeneity at the cellular level. For that, researchers need cell culture and in vivo models where individually labeled cells can be imaged. In a recent article in Cell Reports, researchers have perfected a technique that does precisely that.
Glycolysis in tumors is frequently upregulated, leading to increased biosynthesis of metabolic intermediates required for cell proliferation. Cells can achieve this by upregulation of phosphatidylinositol 3-kinase (PI3K) signaling, either through the acquisition of activating mutations, inactivating mutations in the negative regulatory phosphatase PTEN, or as a result of increased receptor tyrosine kinase signaling. PI3K signaling acts in part through AKT (Protein kinase B) to enhance glycolytic flux, as well as via other mediators that ultimately control the actin skeleton and local membrane dynamics. In estrogen-receptor-positive tumors (ER+), several PI3K inhibitors have now been approved as treatments, which might also be explored for more resistant triple-negative tumor sub-types.
The researchers used a combined approach of glucose fluorescence resonance energy transfer (FRET) imaging, metabolomics, reactive oxygen species detection, and mitochondrial membrane potential fluorescence imaging (with TMRE label) to define tumor heterogeneity at the single cell level. They also used flow cytometry to isolate high- and low-glucose concentration cells for culture for more specific analysis. MCF-7 breast cancer cells were cultured in different kinds of media to reflect different tumor environments, and then cultures were implanted into the mammary fat pad of mice. Implanting tumor cell lines back into the species and tissue of origin (known as orthotopic models) can preserve the normal immune-tumor interactions.
Alternatively, patient-derived xenografts (PDXs) can also be implanted in immunosuppressed mouse models to evaluate human cancers in a defined setting. Traditionally, cancer cells have been implanted subcutaneously for convenient study in mice. However, these tumors prematurely ulcerate, terminating the study prematurely. New assay protocols like subQperior, in which the cells are injected directly into the stroma of the mammary fat pad, have emerged as workhorse tumor models for cancers of all kinds.
The researchers found tumors were interspersed with distinct regions of high-glucose and low-glucose cells that maintained their state for greater than 10 hours. These metabolic states proved to be heritable to daughter cells across mitosis, and even neighboring cells could be radically different. Researchers suspected that the basis for heritable metabolic state could be epigenetic regulation. By testing several inhibitors, they found that while PI3K inhibition knocked down many of the cells, better results were obtained when certain “bromodomain”-containing proteins were simultaneously inhibited. Bromodomains are 110 amino acid motifs that recognize epigenetic acetylation marks on the N-terminal tails of histone proteins. When bromodomain “readers” of lysine acetylation are out of whack, cells can be refractory to PI3K inhibition. At least 43 bromodomain proteins, many histone acetyltransferases, have been identified to date.
Source: Read Full Article